4 Data Wrangling
This week our goals are to be able to:
- Use the dplyr package to perform basic data transformation and analysis
- Filter and arrange rows of datasets
- Create new columns with
mutate - Select columns
- Use pipes to make our code more readable
- Summarize data by groups using
summarize
4.1 Connection to previous work on Data Organization
This week, we will finally see why organized data is worth the effort. We’ll follow an exercise using a data source with over 300,000 rows! The work this week will show us (1) why R is awesome and fast for analysis, and (2) reinforce the purpose of organized data (following the 12 best practices we learned in Week 1).
Because we are dealing with large datasets now, make sure that your Problem Set does not include pages and pages of data by just showing the top of the final result using head(dataset)
4.2 Source
This exercise follows along with the reading for this week R for Data Science Chapter 3 https://r4ds.hadley.nz/data-transform (this was chapter 5 in the old version https://r4ds.had.co.nz/transform.html, hopefully I’ve updated everything but incase I haven’t there’s the link). The template below is for you to be able to follow along in the reading and complete the exercises.
4.3 3.1 Introduction - Example dataset nycflights13
I’ve gone ahead and installed the 2 packages, but you need to load them into the environment using:
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.6
## ✔ forcats 1.0.1 ✔ stringr 1.6.0
## ✔ ggplot2 4.0.1 ✔ tibble 3.3.1
## ✔ lubridate 1.9.4 ✔ tidyr 1.3.2
## ✔ purrr 1.2.1
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsWhy is it important to do this? When you are creating code, explicitly turning on packages that are required is considered good practice. This goes along with the importance of being intentional and making your code reproducible by anyone, anywhere. Tell the computer what to do…explicitly! Tell everyone explicitly what you have done to get to your results.
This also keeps your R sessions memory low and prevents duplicate functions from being loaded from different packages. Notice above when we load tidyverse we get the message that ✖ dplyr::filter() masks stats::filter() and ✖ dplyr::lag() masks stats::lag(), that is because the stats package also has filter and lag functions as well as the dplyr package which is part of the tidyverse package. The tidyverse is actually a package of packages including ggplot2, purrr, tibble, dplyr, tidyr, stringr, readr, and forcats (and maybe more since writing this). We will learn more about all of these in coming weeks. In our case, because we more recently loaded tidyverse if we call filter(some_argument...) this will run the tidyverse/dplyr version of the function. As it says in the reading, if you want to use the base, or stats, version of these functions after loading dplyr, you’ll need to specify the package that the function comes from using two colons :: as in stats::filter() and stats::lag().
4.4 3.1.2
Run flights in the code chunk below. The output should match the reading. Note that you can find a nice README/data dictionary/documentation of this dataset by viewing its help documentation ?flights.
## # A tibble: 336,776 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## 7 2013 1 1 555 600 -5 913 854
## 8 2013 1 1 557 600 -3 709 723
## 9 2013 1 1 557 600 -3 838 846
## 10 2013 1 1 558 600 -2 753 745
## # ℹ 336,766 more rows
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>As described, flights is a data frame called a tibble. What does int mean on the third line of the table? Or dbl?
These are types of variables. Be sure to familiarize yourself with the various types as you move forward, so focus on this section in the reading. You can also check out Vectors and data types in Data Carpentry.
4.5 3.1.3 dplyr basics
https://r4ds.hadley.nz/data-transform#dplyr-basics
Most of the tidyverse aims to make programming make “grammatical” sense in that it is easy to read, understand, and talk about using typical language. One of the really tricky parts of many programming languages including R that you have already experienced is how nested accessors (like [] and $) combined with functions and logical statements are used to do operations on parts of datasets (like finding the mean of certain columns from certain rows). This can make reading a line of code really difficult. You have to read the code from the inside out. For example, from last week, we can run from inside to out.
## [1] 0.3333333## [1] 1 NA 1## [1] FALSE TRUE FALSE## [1] 0.3333333From the dplyr homepage: “dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges” where verbs are functions that operate on nouns, which are your dataset and elements within it.
For all dplyr “verbs”:
The first argument is always a data frame.
The subsequent arguments typically describe which columns to operate on, using the variable names (without quotes).
The output is always a new data frame.
Direct from R4DS: “Because each verb does one thing well, solving complex problems will usually require combining multiple verbs, and we’ll do so with the pipe, |>. We’ll discuss the pipe more in Section 3.4, but in brief, the pipe takes the thing on its left and passes it along to the function on its right so that x |> f(y) is equivalent to f(x, y), and x |> f(y) |> g(z) is equivalent to g(f(x, y), z). The easiest way to pronounce the pipe is “then”. That makes it possible to get a sense of the following code even though you haven’t yet learned the details:
flights |>
filter(dest == "IAH") |>
group_by(year, month, day) |>
summarize(
arr_delay = mean(arr_delay, na.rm = TRUE)
)## `summarise()` has grouped output by 'year', 'month'. You can override using the
## `.groups` argument.## # A tibble: 365 × 4
## # Groups: year, month [12]
## year month day arr_delay
## <int> <int> <int> <dbl>
## 1 2013 1 1 17.8
## 2 2013 1 2 7
## 3 2013 1 3 18.3
## 4 2013 1 4 -3.2
## 5 2013 1 5 20.2
## 6 2013 1 6 9.28
## 7 2013 1 7 -7.74
## 8 2013 1 8 7.79
## 9 2013 1 9 18.1
## 10 2013 1 10 6.68
## # ℹ 355 more rowsdplyr’s verbs are organized into four groups based on what they operate on: rows, columns, groups, or tables. In the following sections you’ll learn the most important verbs for rows, columns, and groups, then we’ll come back to the join verbs that work on tables in Chapter 19. Let’s dive in!”
4.6 3.2 Row-wise functions
4.6.1 3.2.1 filter()
The example filters the data based on month and day. jan1 <- filter(flights, month == 1, day == 1)
The double-equals ==sign implies “is equal to”; in the filter function above, all flights on the first day of January are saved as a new variable jan1.
What is happening in the command below?
## # A tibble: 27,004 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## 7 2013 1 1 555 600 -5 913 854
## 8 2013 1 1 557 600 -3 709 723
## 9 2013 1 1 557 600 -3 838 846
## 10 2013 1 1 558 600 -2 753 745
## # ℹ 26,994 more rows
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>The reading also points out the use of the near function. Why is this important? Illustrate the example below in the code chunk below to reinforce the concept.
Paste sqrt(1.9999999999999999999999)^2 in the code chunk and run it. If you keep removing the trailing 9s, when does the result not equal 2? What happens when you run sqrt(2)^2==2? Show me that you can have the computer make these equivalent using near(), and explain in one word—yes one word—the result of sqrt(2)^2==2 versus using the near function. (Hint: the word starts with P).
4.6.1.1 Logical Operators
We learned about ==, “is equal to,” above. Other logical or Boolean operators that can be used as filters are >, ==, <, <=, != (not equal). You can also combine these with other Logical or Boolean operators: & (and), | (or), and ! (not).
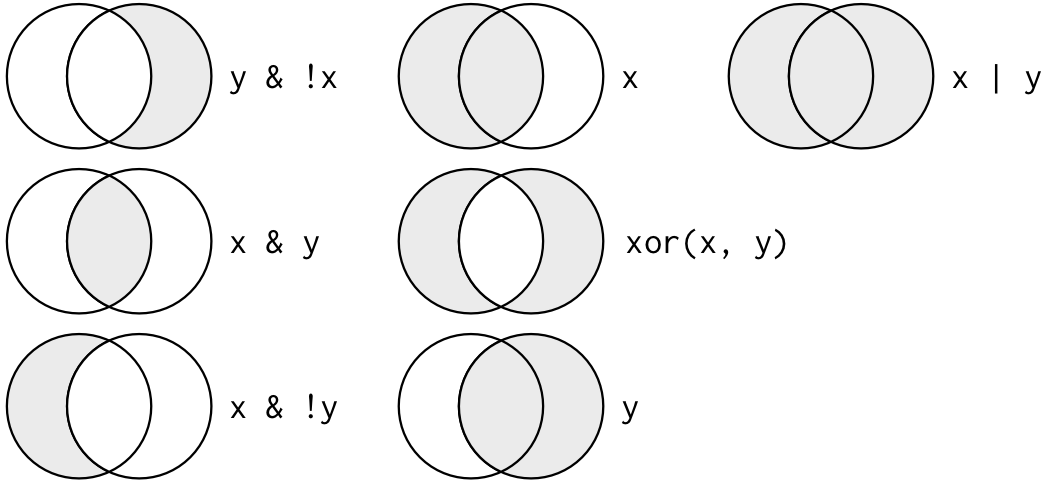
x
## [1] TRUE TRUE TRUE FALSEHow would you select all flights in May and June?
## # A tibble: 57,039 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 5 1 9 1655 434 308 2020
## 2 2013 5 1 451 500 -9 641 640
## 3 2013 5 1 537 540 -3 836 840
## 4 2013 5 1 544 545 -1 818 827
## 5 2013 5 1 548 600 -12 831 854
## 6 2013 5 1 549 600 -11 804 810
## 7 2013 5 1 553 600 -7 700 712
## 8 2013 5 1 553 600 -7 655 701
## 9 2013 5 1 554 600 -6 731 756
## 10 2013 5 1 554 600 -6 707 725
## # ℹ 57,029 more rows
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>## [1] 28796## [1] 28243## [1] 57039## [1] 28520## [1] 57039R also has another nifty logical operator %in%, which searches for a matches of one vector in another and return true for any matching values. So for example:
## [1] TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [25] FALSE TRUESo we could select all flights in May and June using this now.
## # A tibble: 100 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 6 30 2334 1836 298 50 2015
## 2 2013 6 30 2336 2029 187 231 2359
## 3 2013 6 30 2343 2029 194 205 2303
## 4 2013 6 30 2345 2146 119 229 30
## 5 2013 6 30 2347 2125 142 105 2253
## 6 2013 6 30 2348 2130 138 229 14
## 7 2013 6 30 2354 2245 69 53 2359
## 8 2013 6 30 2354 2245 69 117 1
## 9 2013 6 30 2357 2112 165 223 2359
## 10 2013 6 30 2358 2225 93 49 2330
## # ℹ 90 more rows
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>4.6.2 3.2.3 Arranging rows
We can arrange rows by a particular columns values using arrange. For example with the flights dataset we could arrange by departure time.
## # A tibble: 336,776 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 13 1 2249 72 108 2357
## 2 2013 1 31 1 2100 181 124 2225
## 3 2013 11 13 1 2359 2 442 440
## 4 2013 12 16 1 2359 2 447 437
## 5 2013 12 20 1 2359 2 430 440
## 6 2013 12 26 1 2359 2 437 440
## 7 2013 12 30 1 2359 2 441 437
## 8 2013 2 11 1 2100 181 111 2225
## 9 2013 2 24 1 2245 76 121 2354
## 10 2013 3 8 1 2355 6 431 440
## # ℹ 336,766 more rows
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>By default arrange sorts the rows from low to high on the variable you pass. To sort high to low you put a - in front of the variable or use desc(variable).
## # A tibble: 336,776 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 10 30 2400 2359 1 327 337
## 2 2013 11 27 2400 2359 1 515 445
## 3 2013 12 5 2400 2359 1 427 440
## 4 2013 12 9 2400 2359 1 432 440
## 5 2013 12 9 2400 2250 70 59 2356
## 6 2013 12 13 2400 2359 1 432 440
## 7 2013 12 19 2400 2359 1 434 440
## 8 2013 12 29 2400 1700 420 302 2025
## 9 2013 2 7 2400 2359 1 432 436
## 10 2013 2 7 2400 2359 1 443 444
## # ℹ 336,766 more rows
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>This may seem a little awkward, but it allows you to easily provide multiple variable names for a complex sort.
## # A tibble: 336,776 × 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 12 29 2400 1700 420 302 2025
## 2 2013 7 7 2400 1950 250 107 2130
## 3 2013 9 12 2400 2000 240 203 2230
## 4 2013 7 28 2400 2059 181 247 2322
## 5 2013 2 11 2400 2135 145 251 35
## 6 2013 7 17 2400 2142 138 54 2259
## 7 2013 6 17 2400 2145 135 102 2315
## 8 2013 7 13 2400 2155 125 225 43
## 9 2013 7 13 2400 2245 75 101 2359
## 10 2013 8 10 2400 2245 75 110 1
## # ℹ 336,766 more rows
## # ℹ 11 more variables: arr_delay <dbl>, carrier <chr>, flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>4.7 3.3 Columns
4.7.1 3.3.1 Mutate - Add new variables
The function mutate is used to add new variables/columns to a data frame.
Following the example at the beginning of section 5.5 in the book, add a new speed variable using mutate to your data frame.
flights_sml <- select(flights,
year:day,
ends_with("delay"),
distance,
air_time
)
#add a speed variableNext, pay attention to the Useful transformation functions and the modular arithmetic section to obtain hour and minutes from the departure data. Try for yourself below. This is pretty cool and can be useful.
4.7.2 3.3.2 Select
It’s not uncommon to get datasets with hundreds or even thousands of variables. In this case, the first challenge is often narrowing in on the variables you’re actually interested in. select() allows you to rapidly zoom in on a useful subset using operations based on the names of the variables.
select() is not terribly useful with the flights data because we only have 19 variables, but you can still get the general idea:
## # A tibble: 336,776 × 3
## year month day
## <int> <int> <int>
## 1 2013 1 1
## 2 2013 1 1
## 3 2013 1 1
## 4 2013 1 1
## 5 2013 1 1
## 6 2013 1 1
## 7 2013 1 1
## 8 2013 1 1
## 9 2013 1 1
## 10 2013 1 1
## # ℹ 336,766 more rows4.8 3.4 The pipe
The pipe operator, which I’ve demonstrated just a few times above, is really fantastically justified by the The pipe section. I would definitely recommend reading this short section.
The idea of piping is that it can make it easier to write, follow, and understand what the commands are doing. Think of each pipe command as “then”. The pipe command uses the following syntax : |>. What it essentially does is take the result of the code on the left-hand side or previous line(s) and pass it as the first argument to the function on the right-hand side.
We can recreate the example above with pipes. Written in words the code chunk below would be: ASSIGN a new object name,
CHOOSE the dataset to operate on, THEN
arrange the dataset by longest distance, THEN
filter the December flights, THEN
select the flight number, departure delay, and carrier columns.
4.9 3.5 Groups
Grouped summaries are essentially what pivot tables are in Excel, if you have ever heard of those. By using the summarise() function with the group_by function we can, for example find the average flight delay by month. This becomes really awesome! This example starts with using group_by to group the data, then applies summarise.
## # A tibble: 12 × 2
## month delay
## <int> <dbl>
## 1 1 10.0
## 2 2 10.8
## 3 3 13.2
## 4 4 13.9
## 5 5 13.0
## 6 6 20.8
## 7 7 21.7
## 8 8 12.6
## 9 9 6.72
## 10 10 6.24
## 11 11 5.44
## 12 12 16.6Next week we’ll practice summarizing data a lot more as well as “pivoting” our data, which is We’ll get to making those sweet, sweet plots soon.